DNA sequence motifs
When looking at the DNA motif programs, I found that MEME was the most intuitive/easy to use for someone who has limited computer skills, such as myself. I did not like the others because they made it quite hard to do anything with them.
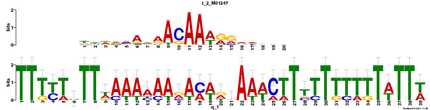
Using MEME, I found that there were 3 motifs found using the human Sox9 genomic DNA sequence, in the hopes of finding cis-regulatory elements and other miscellaneous DNA motifs. Each motif shown below has an opposite strand representation that is not shown. It will be interesting to look into the functional significance of these motifs, if it has not already been done.
In addition to finding these motifs, I used a the TOMTOM application for motif comparison to see what potential transcription factors could bind to the motif. Due to the high amount of potential motif matches, I just chose to represent just the matches that were on the top of the page. If one goes thru MEME to do the TOMTOM themselves, they can see the others (warning: it takes forever to analyze the data).
Using MEME, I found that there were 3 motifs found using the human Sox9 genomic DNA sequence, in the hopes of finding cis-regulatory elements and other miscellaneous DNA motifs. Each motif shown below has an opposite strand representation that is not shown. It will be interesting to look into the functional significance of these motifs, if it has not already been done.
In addition to finding these motifs, I used a the TOMTOM application for motif comparison to see what potential transcription factors could bind to the motif. Due to the high amount of potential motif matches, I just chose to represent just the matches that were on the top of the page. If one goes thru MEME to do the TOMTOM themselves, they can see the others (warning: it takes forever to analyze the data).
Motif 1
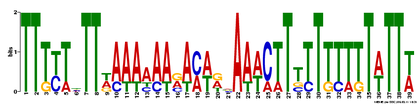
E-value = 8.6e-006 (MEME)
This is by far, for me, is the most exciting potential TF binding site in this motif. Nanog is an essential gene involved in maintaining pluripotency in embryonic stem cells (Mitsui et al 2003). It is known that Nanog is able to maintain chondrocyte differentiation in culture (Zheng et al 2010), but it I do not know if it has been tested to functionally bind to each other. A good experiment would to perform ChIP using an antibody against Nanog, and primers to this motif. It would be interesting to see a functional test of this binding site to see if Nanog truly acts in this fashion.
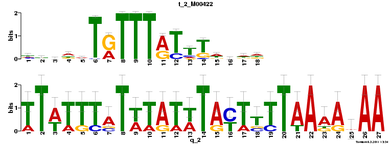
Motif 2

E-value = 3.7e+000 (MEME)
For motif 2, it has been found that Foxj2 has a potential binding site to it. There seems to be no data on Foxj2 interacting with Sox9's cis-regulatory elements, so this would be an interesting avenue to pursue by testing for Foxj2's role in regulating Sox9.
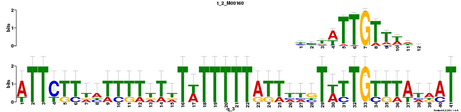
Motif 3
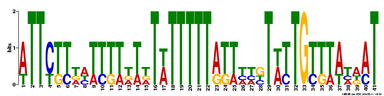
E-value = 1.2e+001 (MEME)
For motif 3, what I found interesting (aside from a huge possibility of other TFs binding to it) was that this is where apparantly the SRY protein binds to. This is of interest because SRY is known as the master regulator of male sex determination in embryonic development (Sinclair et al 1990). This is not surprising because Sox9 mutants often have a sex reversal phenotype (Wagner et al. 1994).
References
Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003 May 30;113(5):631-42.
Zheng H, Gourronc F, Buckwalter JA, Martin JA. Nanog maintains human chondrocyte phenotype and function in vitro. J Orthop Res. 2010 Apr;28(4):516-21
Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990 Jul 19;346(6281):240-4.
Wagner, T., Wirth, J., Meyer, J., Zabel, B., Held, M., Zimmer, J., Pasantes, J., Dagna Bricarelli, F., Keutel, J., Hustert, E., Wolf, U., Tommerup, N., Schempp, W., Scherer, G. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79: 1111-1120, 1994.
Zheng H, Gourronc F, Buckwalter JA, Martin JA. Nanog maintains human chondrocyte phenotype and function in vitro. J Orthop Res. 2010 Apr;28(4):516-21
Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990 Jul 19;346(6281):240-4.
Wagner, T., Wirth, J., Meyer, J., Zabel, B., Held, M., Zimmer, J., Pasantes, J., Dagna Bricarelli, F., Keutel, J., Hustert, E., Wolf, U., Tommerup, N., Schempp, W., Scherer, G. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79: 1111-1120, 1994.