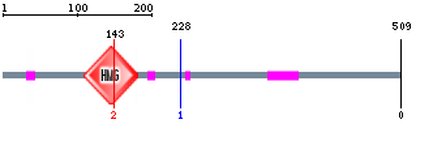
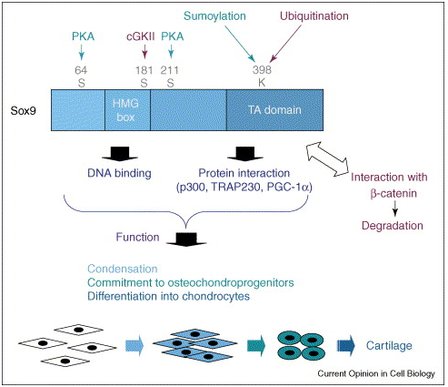
Sox9 protein structure and domains
Going off from the SMART database analysis of Sox9, there was little recognizable protein motifs except for the HMG box domain. The HMG (High Mobility Group) box is defined as a DNA-binding domain that induces severe bends in the target DNA, but has little or no sequence specificity. This domain was the one domain that always showed up in all of the protein domain finding programs used in this study.
Interestingly enough, according to SMART, there are numerous low complexity regions, which suggests as domains of some sort. Most likely interaction domains of the sort. However, the functional significance, if they exist at all as an actual domain, is unclear.
The pfam site's algorithm shows an additional domain known as the SoxN domain, which contains the dimerization domain in Sox9. This one seems to be specific to the Sox family of proteins, and is in the same position in other Sox9 homologs, which indicates conservation of the general superstructure in at least those regions (even though a good verification would be to generate a crystal structure of Sox9 and its homologs). Unfortunately, none of the programs were able to identify the transactivation domain of Sox9, which is described in the literature (Kawakami et al 2006). This may be attributed to the fact that the TA domain is not highly conserved in other proteins.
Interestingly enough, according to SMART, there are numerous low complexity regions, which suggests as domains of some sort. Most likely interaction domains of the sort. However, the functional significance, if they exist at all as an actual domain, is unclear.
The pfam site's algorithm shows an additional domain known as the SoxN domain, which contains the dimerization domain in Sox9. This one seems to be specific to the Sox family of proteins, and is in the same position in other Sox9 homologs, which indicates conservation of the general superstructure in at least those regions (even though a good verification would be to generate a crystal structure of Sox9 and its homologs). Unfortunately, none of the programs were able to identify the transactivation domain of Sox9, which is described in the literature (Kawakami et al 2006). This may be attributed to the fact that the TA domain is not highly conserved in other proteins.
HMG box description: The HMG (High Mobility group) box is a DNA-binding motif that induces severe bends in the target DNA but has little or no sequence specificity.
Tertiary structure of Sox9
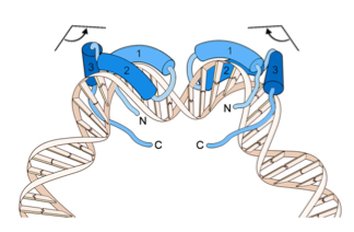
Here is a cartoon of the tertiary structure of Sox9 interacting with DNA, and subsequently bending it (Lefebvre et al 2007). Notice how the HMG box is the domain that is interacting with the DNA and it is physically bending the strand. Sox9's chondrocyte-specific DNA binding activity is dependent on Sox9 dimerizing with itself via the dimerization domain (Coustry et al 2010; Bernard et al 2003) found within the SoxN domain of the protein.
Molecular surfaces of Sox9 HMG domain interacting with DNA d(GCACAAAC)
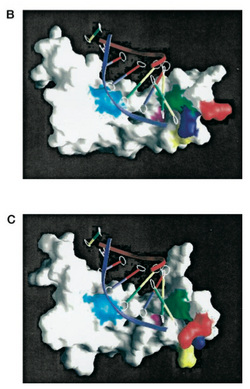
This image to the left (taken from McDowall et al 1999) further demonstrates the fact that Sox9 binds to DNA and the structure of the HMG box is essential for proper DNA-Protein interactions.
In the figure:
B) is from wildtype Sox9. C) is from P70R mutant Sox9, which causes a conformational change in the HMG domain, which affects its ability to bind to the selected DNA sequence. These figures are from McDowall et al 1999.
Other Biochemical Information on Sox9
Protein Isoelectric Point: pH 6.32
Protein Molecular Weight: 56.15 kD
Protein Molecular Weight: 56.15 kD
Scansite predicted motifs/domains
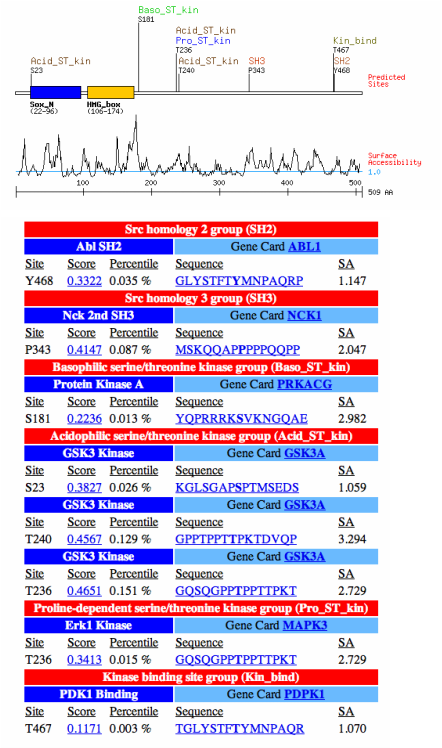
In addition to the predicted Sox_N and HMG_box domains, scansite predicted (with highest constraints) other possible domains that may act as the trans-activation domain for Sox9. These are shown on the figures to the left. Out of all of the programs used to predict protein/domain structure, I preferred Scansite due to it giving the most information, and it also predicted potential post-translational modifications, SH2/3 binding sites, and even surface accessibility based on calculations using the amino acid sequence given. Therefore I would advise anybody looking for in-depth protein domain information and predictions that go beyond simple large domains should go strait to this site to get the most valuable protein information.
References
Yasuhiko
Kawakami, Joaquín Rodriguez-León and Juan Carlos Izpisúa Belmonte. The
role of TGFβs and Sox9 during limb chondrogenesis. Current
Opinion in Cell Biology. Volume 18,
Issue 6, December 2006, 723-729
Lefebvre V, Dumitriu B, Penzo-Méndez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39(12):2195-214.
Coustry F, Oh CD, Hattori T, Maity SN, de Crombrugghe B, Yasuda H. (2010). The dimerization domain of SOX9 is required for transcription activation of a chondrocyte-specific chromatin-DNA template. Nucleic Acids Res. 38, 6018-6028
Bernard P, Tang P, Liu S, Dewing P, Harley VR, Vilain E. (2003). Dimerization of SOX9 is required for chondrogenesis, but not for sex determination. Hum Mol Genet. 12, 1755-1765
McDowall S, Argentaro A, Ranganathan S, Weller P, Mertin S, Mansour S, Tolmie J, Harley V. (1999)
Functional and structural studies of wild type SOX9 and mutations causing campomelic dysplasia. J Biol Chem. 274(34):24023-30.
Lefebvre V, Dumitriu B, Penzo-Méndez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39(12):2195-214.
Coustry F, Oh CD, Hattori T, Maity SN, de Crombrugghe B, Yasuda H. (2010). The dimerization domain of SOX9 is required for transcription activation of a chondrocyte-specific chromatin-DNA template. Nucleic Acids Res. 38, 6018-6028
Bernard P, Tang P, Liu S, Dewing P, Harley VR, Vilain E. (2003). Dimerization of SOX9 is required for chondrogenesis, but not for sex determination. Hum Mol Genet. 12, 1755-1765
McDowall S, Argentaro A, Ranganathan S, Weller P, Mertin S, Mansour S, Tolmie J, Harley V. (1999)
Functional and structural studies of wild type SOX9 and mutations causing campomelic dysplasia. J Biol Chem. 274(34):24023-30.