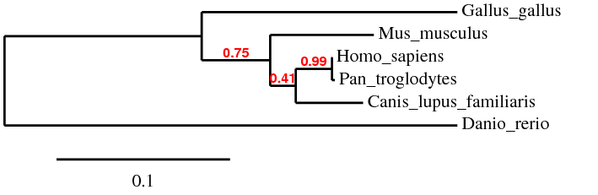
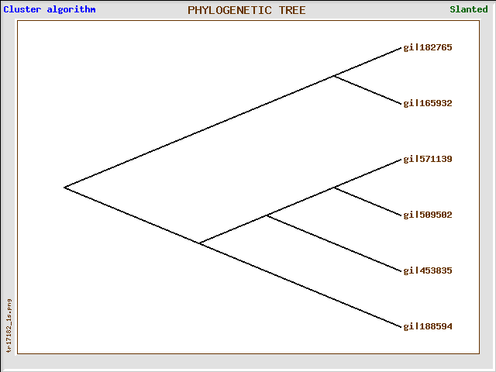
Phylogeny (nucleotide)
When I used the phylogeny programs for my nucleotide sequences, I decided to use mRNA/cDNA sequences available to me instead of genomic DNA. I did this because introns and other non-coding sequences are not expected to be well conserved, which would could possibly skew what the phylogeny is. Coding sequence is under higher selection (nucleotide less than amino acid, however) than non-coding sequences, typically. With the tree generated by MABL, the general organization to the tree is not really all that surprising. The only thing of note is that the branch-point between the primate and canine lineages has a bootstrap value less than .50, which indicates little confidence there. Most programs would get collapse that into the previous branch. I hypothesize that these bootstraps are smaller than the protein because synonymous mutations in the coding region would be detected in the mRNA, but will pass by unnoticed in the protein sequence.
Looking at the Gene Bee tree, which is oddly organized for reasons unknown to me, shows generally that the same pattern of divergence is shown. Of course, this tree did not undergo bootstrapping, the data represented is not very strongly supported. I have to admit that the MABL program is much better than the Gene Bee program because of its ease of use, and is much easier to interpret. The Gene Bee tree just shows gi numbers, which can be confusing and cumbersome to look up the gi's all the time.